Key tools developed by our project described below. Click on each to learn more!!
|
ECOTOXXPLORER.CA
EcoToxXplorer.ca provides intuitive bioinformatics support for users to analyze EcoToxChip results as well as transcriptomics (e.g., RNA-seq) data. It has been modeled on our successful cloud-based tools (metaboanalyst.ca; networkanalyst.ca). Try it out!! Go to the website, click on "RNAseq" (to the left), and then scroll down to "try our test data". |
|
FASTBMD.CA
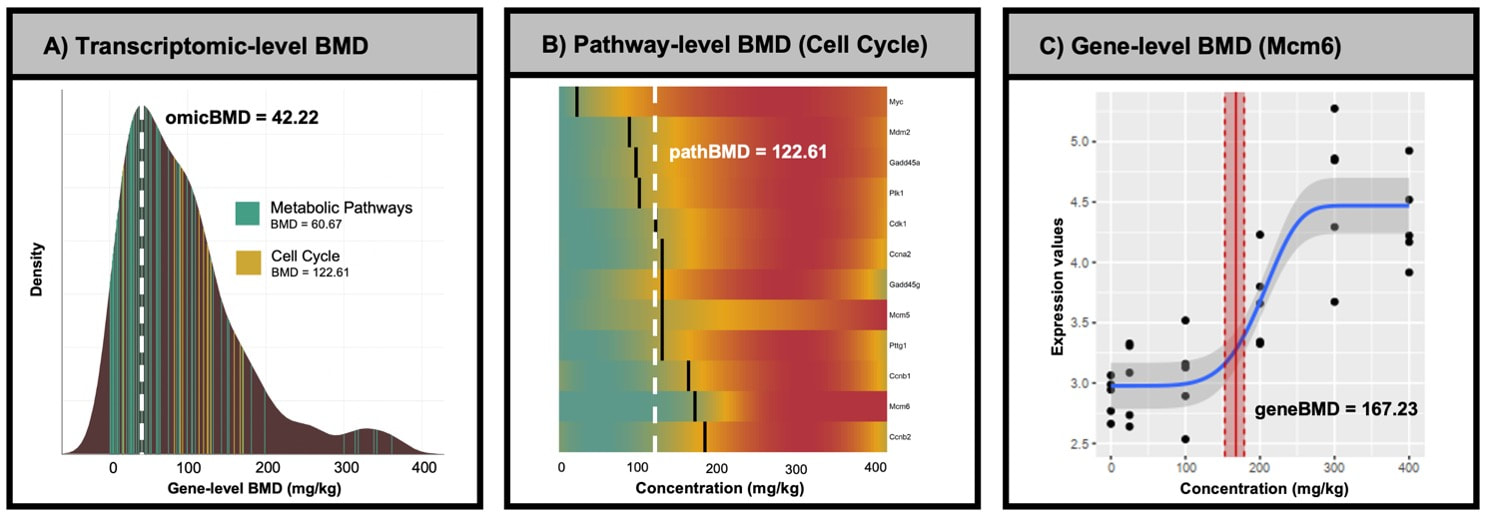
fastBMD.ca is a web-based tool that implements standardized methods for transcriptomics benchmark dose–response analysis in R. It is >60 times faster than the current leading software, supports transcriptomics data from 13 species, and offers a comprehensive analytical pipeline that goes from processing and normalization of raw gene expression values to interactive exploration of pathway-level benchmark dose results. |
A TIMELINE OF THE DEVELOPMENT OF NEW APPROACH METHODOLOGIES
|
Timeline content research led by Matthieu Mondou
for the GE3LS research component. |
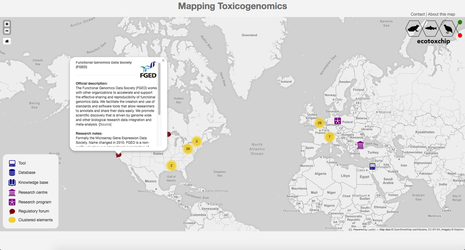
Mapping Toxicogenomics
|
Map design and content research led by Matthieu Mondou
Map development by Tássia Camões Araujo for the GE3LS research component. |